Abstract
Epigenetic modifications are critical determinants of cell identity and play a central role in normal hematopoietic differentiation. Moreover, mutations in epigenetic modifiers including TET2 and IDH2 mutations, which result in a decreased ability to de-methylate DNA, are frequent in myeloid leukemias and clonal hematopoiesis of indeterminate potential. This raises a central question in normal and pathologic hematopoiesis - how do epigenetic modifiers shape the differentiation landscape?
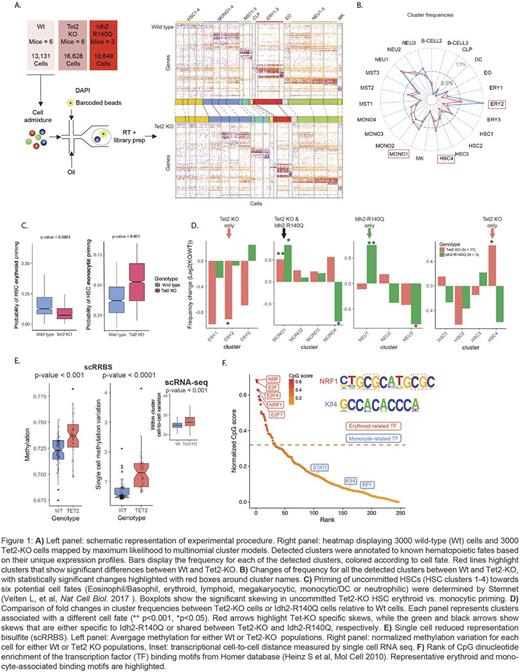
To address this question, we applied high-throughput single-cell RNA-seq (scRNAseq) to reconstruct high-resolution maps of hematopoietic differentiation, with and without disruption of central epigenetic modifiers (Tet2 and Idh2). Briefly, FACS-sorted lineage-negative bone marrow cells were isolated from sixteen-week old Mx1 -cre wildtype (Wt, n=6), Mx1 -cre Tet2fl/fl (n= 6) or Mx1 -cre Idh2R140Q/Wt (n=3) mice, 4 weeks following recombination. In total, 45,100 cells underwent droplet-based scRNAseq. Clustering of Wt cells identified 25 distinct progenitor populations, recapitulating previous reports and providing a map of normal mouse hematopoietic differentiation.
Tet2knockout (KO) resulted in a 1.5-fold increase in uncommitted cluster HSC4 (Flt3hi, c-Kithi, Cd34low) (Figure A-B), consistent with a potential differentiation block resulting from Tet2-KO. Notably, the HSC differentiation block did not uniformly affect differentiation into committed progenitors, but resulted in a 2-fold decrease in erythroid precursors and a 1.5-fold increase in monocyte precursors. These findings were evident by scRNA-seq four months before this phenotype was observed by flow cytometry. While the expression profiles of these committed erythroid and monocytic progenitors were not different from Wt progenitors, the degree of priming to these fates within uncommitted Tet2-KO HSCs (HSC clusters 1-4) showed reduced priming towards the erythroid fate and increased priming towards the monocytic fate (Figure C). In contrast, mutant Idh2 resulted in different skews of hematopoiesis than Tet2-KO, including an increase in neutrophil precursors, while there was no significant change in the frequency of erythroid precursors (Figure D). These data demonstrate that Tet2 and Idh2 mutations, both of which disrupt de-methylation, have distinct effects on hematopoietic differentiation.
To explore the link between aberrant DNA methylation upon Tet2 - KO and the observed differentiation skews, we performed joint single-cell methylation and RNAseq, as well as ATAC-bisulfite sequencing. As expected, Tet2-KO resulted in higher global methylation. However, the increase in methylation was stochastically distributed across the genome, resulting in higher methylation heterogeneity (Figure E), and associated with higher cell-to-cell variation in transcriptional profiles. Importantly, we found that stochastic methylation changes result in deterministic differentiation skews due to differential sensitivity to methylation of binding motifs of lineage-defining transcription factors (TFs). For instance, motifs of erythroid-related TFs (including Nrf1, E2f4 and E2f1), but not monocytic-related TFs, showed a marked enrichment of CpGs and methylation-sensitive binding (Figure F).
These data demonstrate the ability of high-resolution single-cell mapping of hematopoiesis to identify the exact point of disruption of HSC differentiation due to alterations in epigenetic modifiers. Specifically, these data show that Tet2-KO results in reduced erythroid but increased monocytic priming of HSCs, leading to skewed differentiation. Moreover, single-cell epigenomic analyses identified that while the hypermethylation effect of Tet2-KO is distributed widely across the genome, the skews in lineage priming result from biases in the sensitivity of lineage-defining TFs to CpG methylation at their motifs. The skews may therefore arise not from a change in methylation of a particular locus, but rather through broad lack of demethylation in targets of erythroid priming TFs, resulting from inherent enrichment of these motifs in CpGs.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal